Current Research
Investigation of the molecular basis of heterosis by precision proteomics
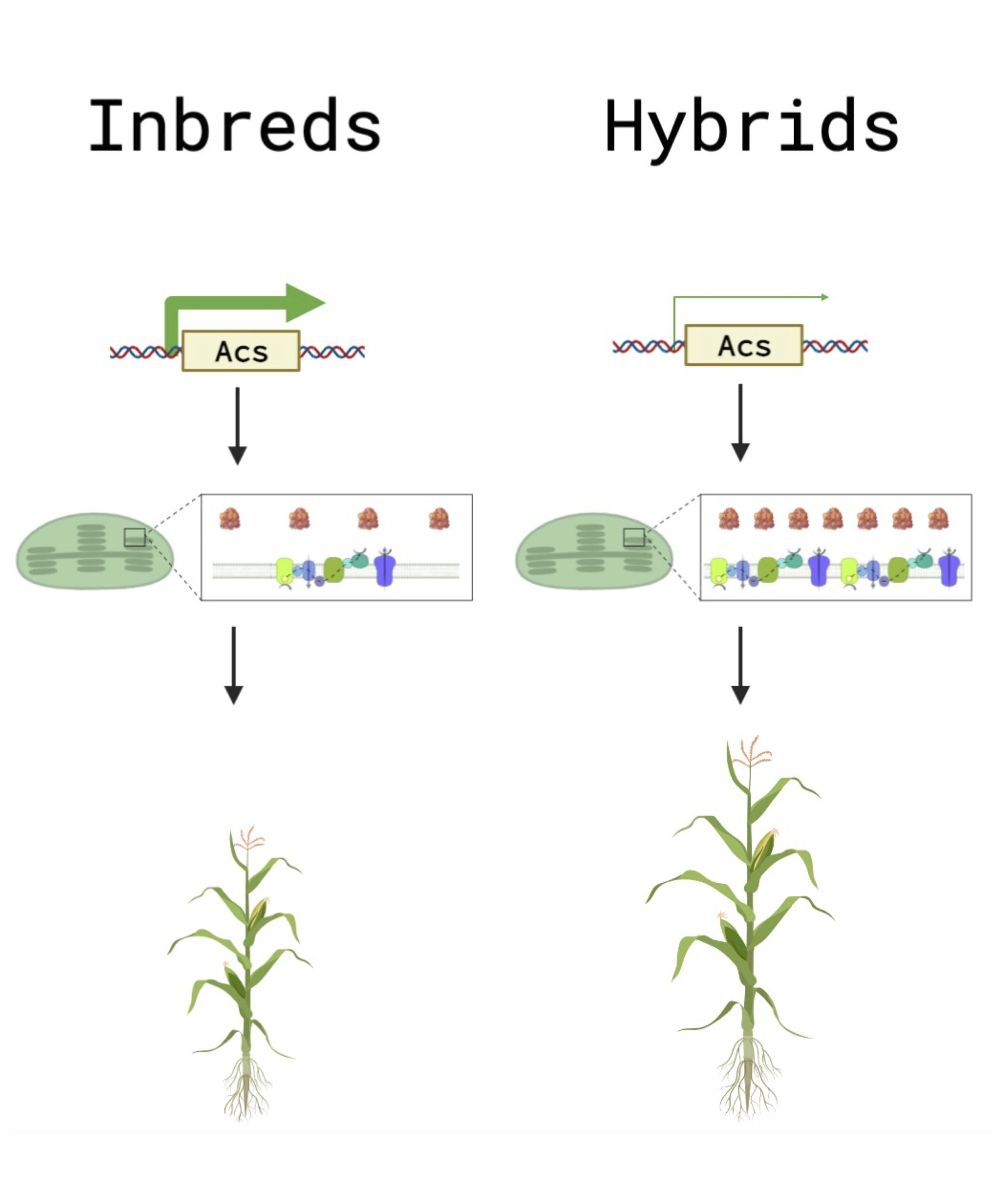
Despite the importance of heterosis in agriculture, its molecular underpinnings have persisted as an unsolved classical problem in biology since its initial report by Charles Darwin. There are few instances of gene expression outside mid-parental range in hybrids, suggesting a post-transcriptional mechanism for hybrid vigor. We employ precision proteomics to analyze the proteomes of inbred plants and their F1 hybrids. We have identified a novel molecular phenotype of hybrids; the abundance of plastid photosynthetic protein complexes, consisting of subunits encoded by both the nucleus and the plastid, was elevated in the hybrid relative to mid-parent levels, which may account for the greater photosynthetic capacity of hybrids. This pattern was not reflected in RNA-seq data. Furthermore, we have identified a striking positive correlation between expression heterosis (hybrid/mid-parent expression levels) of the plastid ribosome and plant height heterosis (hybrid/mid-parent plant height). Additionally, we have found that ethylene biosynthetic enzymes were expressed below mid-parental levels in the hybrid, and an ethylene biosynthesis mutant, acs2/6, partially phenocopied the proteome of the hybrid, indicating that a reduction in ethylene biosynthesis may be upstream of part of the hybrid molecular phenotype.
Past Research
Integration of omic networks in a developmental atlas of maize
Expression of a given gene at the RNA level does not always correlate with expression at the protein level for many organisms. Walley et al. have built an integrated atlas of gene expression and regulatory networks in developing maize, using the same tissue samples to measure the transcriptome, proteome, and phosphoproteome. Coexpression networks from the transcriptome and proteome showed little overlap with each other, even though they showed enrichment of similar pathways. Integration of mRNA, protein, and phosphoprotein datasets improved the predictive power of the gene regulatory networks.
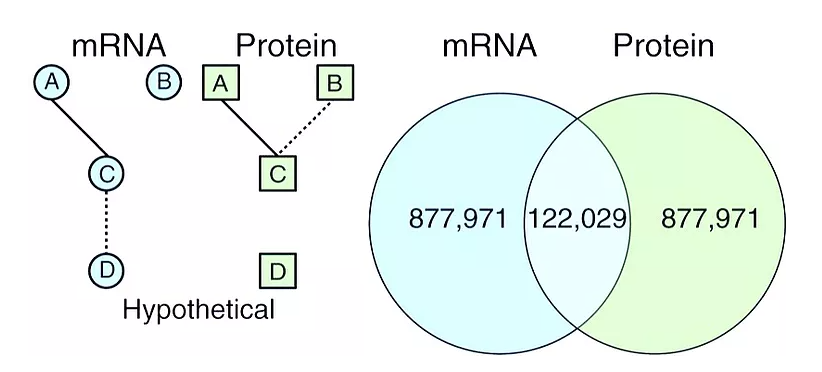
Identification of the expressome by machine learning on omics data
Accurate annotation of plant genomes remains complex due to the presence of many pseudogenes arising from whole-genome duplication-generated redundancy or the capture and movement of gene fragments by transposable elements. Our new method uses only epigenomic patterns to classify the expression potential of annotated genes and identifies pseudogenes that are difficult to classify based solely on sequence. Genes were divided into those with protein expression, those with mRNA expression, and those that are silent. A large fraction of annotated genes are constitutively silent in one lineage but can be transcribed in others. We refer to the species-wide set of transcribed genes as the expressome and show that it is much larger than the expressible gene set in any individual. Additionally, we find that DNA methylation patterns within the gene body can differentiate between genes that express proteins and genes that express only RNAs.